
So, while I answered your question, I'm not sure it's any help at all. Sadly, neither this way I could achieve better performance. We divide x into two halves, then call permMTX in each, then recombine with cbind. So, what if we tried to allocate as many calls to sample sequentially to a single process? This is what I tried here: parPermMTX2 = function(x, cluster) do.call(cbind, parLapply(cl = cluster, X = list(x, x), fun = permMTX)) Perhaps you're running out of memory, and using the hard disk cache, which is really slow. On my system, sampling 68E3 integers 32E3 times takes roughly 40 seconds: microbenchmark(sample(68E3), times = 32E3) See Why is the parallel package slower than just using apply?
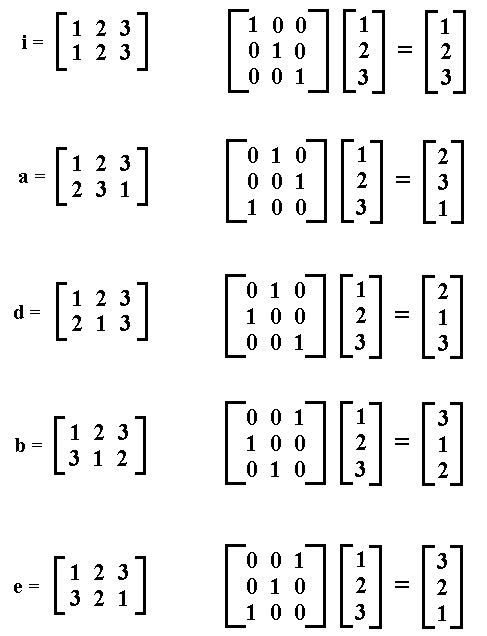
This is due to the fact sample is heavily optimized already, so the overhead of spawning processes is bigger than simply calling sample. While it does run in parallel on my end, it's not faster at all than simply employing apply, but I couldn't test with data at the same scale as yours, so I can't be sure it'll work. I think it's recommended to export the data to the processes as well, you can do that simply calling clusterExport(cl, varlist = "exampleData") Permutation of indices for subscripts In contrast, if you permute a matrix by using subscript indices, no floating-point operations are needed. The way parallel works (spawning multiple R processes) you have to assure that you have enough memory to fit multiple copies of your data as well. To permute the columns, the permutation matrix is p x p, and the matrix multiplication requires O ( Np 2) operations.

Here's an example: cl = makeCluster(detectCores(logical = FALSE)) With parallel you have to register a cluster before usage. ParPermMTX = function(x, cluster) parApply(cl = cluster, X = x, MARGIN = 2L, FUN = sample) Then we can use library parallel to parallelize that function: library(parallel) permMTX = function(x) apply(x, 2L, sample) First, I'd make use of vectorization, which should make it more efficient.


 0 kommentar(er)
0 kommentar(er)
